About
CRISPResso is a software pipeline designed to enable rapid and intuitive interpretation of genome editing experiments. A limited web implementation is available at: http://crispresso2.pinellolab.org/ or http://crispresso.com.
Briefly, CRISPResso:
- Aligns sequencing reads to a reference sequence
- Quantifies insertions, mutations and deletions to determine whether a read is modified or unmodified by genome editing
- Summarizes editing results in intuitive plots and datasets
Tools
CRISPResso is a suite of complementary tools:
- CRISPResso - for analyzing and interpreting single experimental conditions on a single amplicon
- CRISPRessoBatch - for analyzing and comparing multiple experimental conditions at the same site
- CRISPRessoPooled - for analyzing multiple amplicons from a pooled amplicon sequencing experiment
- CRISPRessoWGS - for analyzing specific sites in whole-genome sequencing samples
- CRISPRessoCompare - for comparing editing between two samples (e.g., treated vs control)
- CRISPRessoAggregate - for aggregating results from previously-run CRISPResso analyses
How can you use CRISPResso?
CRISPResso can be used to analyze genome editing outcomes using cleaving nucleases (e.g. Cas9 or Cpf1) or noncleaving nucleases (e.g. base editors). The following operations can be automatically performed:
- Filtering of low-quality reads
- Adapter trimming
- Alignment of reads to one or multiple reference sequences (in the case of multiple alleles)
- Quantification of HDR and NHEJ outcomes (if the HDR sequence is provided)
- Quantification frameshift/inframe mutations and identification affected splice sites (if an exon sequence is provided)
- Visualization of the indel distribution and position (for cleaving nucleases)
- Visualization of distribution and position of substitutions (for base editors)
- Visualization of alleles and their frequencies
CRISPResso processing
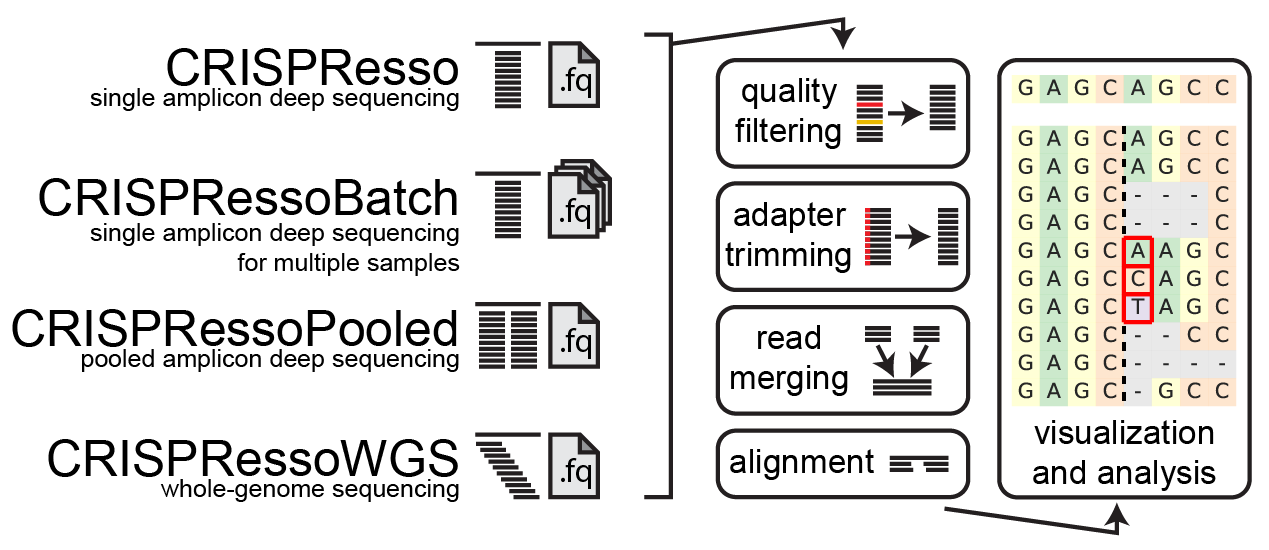
Quality filtering
Input reads are first filtered based on the quality score (phred33) in order to remove potentially false positive indels. The filtering based on the phred33 quality score can be modulated by adjusting the optimal parameters (see additional notes below).
Adapter trimming
Next, adapters are trimmed from the reads. If no adapter are present, select 'No Trimming' under the 'Trimming adapter' heading in the optional parameters. If reads contain adapter sequences that need to be trimmed, select the adapters used for trimming under the ‘Trimming adapter’ heading in the optional parameters. Possible adapters include Nextera PE, TruSeq3 PE, TruSeq3 SE, TruSeq2 PE, and TruSeq2 SE. The adapters are trimmed from the reads using fastp.
Read merging
If paired-end reads are provided, reads are merged using fastp. This produces a single read for alignment to the amplicon sequence, and reduces sequencing errors that may be present at the end of sequencing reads.
Alignment
The preprocessed reads are then aligned to the reference sequence with a global sequence alignment algorithm that takes into account our biological knowledge of nuclease function. If multiple alleles are present at the editing site, each allele can be passed to CRISPResso and sequenced reads will be assigned to the reference sequence or origin.
Visualization and analysis
Finally, after analyzing the aligned reads, a set of informative graphs are generated, allowing for the quantification and visualization of the position and type of outcomes within the amplicon sequence.
How is CRISPResso2 different from CRISPResso?
CRISPResso2 introduces four key innovations for the analysis of genome editing data:
- Comprehensive analysis of sequencing data from base editors. We have added additional analysis and visualization capabilities especially for experiments using base editors.
- Allele specific quantification of heterozygous references. If the targeted editing region has more than one allele, reads arising from each allele can be deconvoluted.
- A novel biologically-informed alignment algorithm. This algorithm incorporates knowledge about the mutations produced by gene editing tools to create more biologically-likely alignments.
- Ultra-fast processing time.
Installation
CRISPResso can be installed using the conda package manager Bioconda, or it can be run using the Docker containerization system.
Bioconda
To install CRISPResso using Bioconda, download and install Anaconda Python, following the instructions at: https://docs.anaconda.com/free/anaconda/install/.
Open a terminal and type:
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
To install CRISPResso into the current conda environment, type:
conda install crispresso2
Alternatively, to create a new environment named crispresso2_env with CRISPResso, type:
conda create -n crispresso2_env -c bioconda crispresso2
Activate your conda environment:
conda activate crispresso2_env
Verify that CRISPResso is installed using the command:
CRISPResso -h
Bioconda for Apple Silicon
If you would like to install CRISPResso using bioconda on a Mac with Apple silicon (aren't sure?), then there is a slight change you need to make. First, ensure that you have Rosetta installed. Next, you must tell bioconda to install the Intel versions of the packages. If you would like to do this system wide, which we recommend, run the command:
conda config --add subdirs osx-64
Then you can proceed with the installation instructions above.
If you would like to use the Intel versions in a single environment, then run:
CONDA_SUBDIR=osx-64 conda create -n crispresso2_env -c bioconda crispresso2
If you choose to use the CONDA_SUBDIR=osx-64 method, note that if you install additional packages into the environment you will need to add the CONDA_SUBDIR=osx-64 to the beginning of each command. Alternatively, you could set this environment variable in your shell, but we recommend to use the conda config --add subdirs osx-64 method because it is less error prone.
Docker
CRISPResso can be used via the Docker containerization system. This system allows CRISPResso to run on your system without configuring and installing additional packages. To run CRISPResso, first download and install docker: https://docs.docker.com/engine/installation/.
Next, Docker must be configured to access your hard drive and to run with sufficient memory. These parameters can be found in the Docker settings menu. To allow Docker to access your hard drive, select 'Shared Drives' and make sure your drive name is selected. To adjust the memory allocation, select the 'Advanced' tab and allocate at least 4G of memory.
To run CRISPResso, make sure Docker is running, then open a command prompt (Mac) or Powershell (Windows). Change directories to the location where your data is, and run the following command:
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso -h
The first time you run this command, it will download the Docker image. The -v parameter mounts the current directory to be accessible by CRISPResso, and the -w parameter sets the CRISPResso working directory. As long as you are running the command from the directory containing your data, you should not change the Docker -v or -w parameters.
Additional parameters for CRISPResso as described below can be added to this command. For example,
docker run -v ${PWD}:/DATA -w /DATA -i pinellolab/crispresso2 CRISPResso -r1 sample.fastq.gz -a ATTAACCAAG
Troubleshooting
Please check that your input file(s) are in FASTQ format (compressed fastq.gz also accepted).
If you get an empty report, please double check that your amplicon sequence is correct and in the correct orientation. It can be helpful to inspect the first few lines of your FASTQ file - the start of the amplicon sequence should match the start of your sequences. If not, check to see if the files are trimmed (see point below).
It is important to determine whether your reads are trimmed or not. CRISPResso2 assumes that the reads ARE ALREADY TRIMMED! If reads are not already trimmed, select the adapters used for trimming under the ‘Trimming Adapter’ heading under the ‘Optional Parameters’. This is FUNDAMENTAL to CRISPResso analysis. Failure to trim adaptors may result in false positives. This will result in a report where you will observe an unrealistic 100% modified alleles and a sharp peak at the edges of the reference amplicon in figure 4.
The quality filter assumes that your reads uses the Phred33 scale, and it should be adjusted for each user’s specific application. A reasonable value for this parameter is 30.
If your amplicon sequence is longer than your sequenced read length, the R1 and R2 reads should overlap by at least 10bp. For example, if you sequence using 150bp reads, the maximum amplicon length should be 290 bp.
Especially in repetitive regions, multiple alignments may have the best score. If you want to investigate alternate best-scoring alignments, you can view all alignments using this tool: http://rna.informatik.uni-freiburg.de/Teaching/index.jsp?toolName=Gotoh. As input, sequences from the 'Alleles_frequency_table.txt' can be used. Specifically, for a given row, the value in the 'Aligned_Sequence' should be entered into the 'Sequence a' box after removing any dashes, and the value in the 'Reference_Sequence' should be entered into the 'Sequence b' box after removing any dashes. The alternate alignments can be selected in the 'Results' panel in the Output section.
License
CRISPResso2 is made available for free to academic researchers under this limited license for non-commercial use.
IMPORTANT: If you plan to use the CRISPResso2 for-profit, you will need to purchase a license. Please contact licensing@edilytics.com for more information.
CRISPResso2 END USER LICENSE AGREEMENT
BEFORE PROCEEDING, PLEASE READ THE END USER LICENSE AGREEMENT BELOW.
BY USING THIS SOFTWARE TOOL YOU ATTEST TO (I) BEING AN ACADEMIC RESEARCHER, (II) USING IT SOLELY FOR RESEARCH PURPOSES AND (III) YOUR ACCEPTANCE OF THE END USER LICENSE AGREEMENT.
-
General. As used herein, the term “you” or “your” means any individual or entity accessing this site or using the software tool “CRISPResso2” (the “Software Tool”) pursuant to this End-User License Agreement (“EULA”).
-
License to Use. The Software Tool is free for your use subject to the terms and conditions set forth below. The General Hospital Corporation, dba Massachusetts General Hospital (“MGH”) reserves the right to change, from time to time and at its sole discretion, this EULA. Your continued use of the Software Tool after any such modification constitutes your agreement and acceptance of such changes.
MGH owns all right, title and interest in the Software Tool. MGH grants to you, the “Licensee,” a royalty-free, non-exclusive, non-transferable, revocable license to use the Software Tool for non-commercial research or academic purposes only; it is NOT made available here as a free tool or download for any commercial or clinical use. You may not copy or distribute the Software Tool in any form. This license is limited to the individual that accesses the Software Tool. No right to sublicense or assign this EULA is granted herein.
The Software Tool optionally makes calls to unmodified versions of fastp https://github.com/OpenGene/fastp software, which is covered under its own license (MIT).
By using this Software Tool, you agree to allow MGH the right to collect data and statistics (i) on system usage patterns and (ii) to improve this Software Tool.
-
Limitations on Use. THE SOFTWARE TOOL HAS NOT BEEN REGISTERED OR APPROVED BY THE U.S. FOOD AND DRUG AGENCY, OR ANY OTHER GOVERNMENTAL AGENCY. THE SOFTWARE TOOL MAY BE USED ONLY AS A REFERENCE TOOL AND FOR CLINICAL EDUCATION, SIMILAR TO THE USE OF A TEXTBOOK OR A JOURNAL ARTICLE. THE SOFTWARE TOOL SHALL NOT BE USED AS A DIAGNOSTIC DECISION MAKING SYSTEM AND MUST NOT BE USED TO MAKE A CLINICAL DIAGNOSIS OR REPLACE OR OVERRULE A LICENSED HEALTH CARE PROFESSIONAL'S JUDGMENT OR CLINICAL DIAGNOSIS.
-
Disclaimer of Warranties. TO THE FULLEST EXTENT PERMITTED BY LAW, MGH PROVIDES THE SOFTWARE TOOL "AS IS" AND “AS AVAILABLE” WITH ALL FAULTS, ERRORS AND DEFECTS, AND NEITHER MGH NOR ANY OF ITS PERSONNEL NOR ANY OF ITS AFFILIATES IS RESPONSIBLE FOR ENSURING THAT ANY USE OF SOFTWARE TOOL WILL BE CLINICALLY SOUND, WITHOUT ERROR, UNINTERRUPTED OR OTHERWISE SUCCESSFUL. THE RIGHTS GRANTED IN THIS EULA ARE MADE AVAILABLE WITHOUT WARRANTY OF ANY KIND, EITHER EXPRESSED OR IMPLIED, INCLUDING, BUT NOT LIMITED TO, IMPLIED WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE, TITLE AND NON-INFRINGEMENT.
-
Limitation of Liability. TO THE FULLEST EXTENT PERMITTED BY LAW, MGH SHALL NOT BE LIABLE TO YOU FOR ANY INDIRECT, INCIDENTAL, SPECIAL OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT WITHOUT LIMITATION, ANY DAMAGES RESULTING FROM LOSS OF USE OR LOST BUSINESS, REVENUE, PROFITS, DATA OR GOODWILL) ARISING IN CONNECTION WITH YOUR USE OF THE SOFTWARE TOOL OR OTHERWISE, WHETHER IN AN ACTION IN CONTRACT, TORT, STRICT LIABILITY, NEGLIGENCE OR OTHERWISE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGES.
-
Indemnification. You agree to defend, indemnify and hold harmless MGH and its affiliates, trustees, officers, employees, staff members, agents or contractors from and against any claim, charge, demand, action or suit, whether in contract, tort, strict liability, negligence or otherwise, for any and all losses, costs, charges, claims, demands, fees, expenses or damages of any nature or kind arising out of, connected with or resulting from (i) the use of the Software Tool by you, your affiliates, employees, staff, faculty, students, agents or (ii) relating in any way to this EULA.
In consideration of MGH providing access to the Software Tool free of charge, you agree not to bring any claim, lawsuit, or action (“Claim”) for any damages, costs, liabilities, settlement amounts and/or expenses (including attorneys’ fees) against MGH or its affiliates, trustees, officers, employees, staff members, agents or contractors arising out of or related to your use of the Software Tool.
- No Other Rights. You do not have the right to use the name, trademark, service mark, logo or other identifying characteristics of MGH or any of its affiliates or employees. All rights not expressly granted herein are reserved by MGH.
MGH may terminate your access to and use of the Software Tool at any time, with or without notice, for any reason or for no reason at all.
-
Governing Law. The construction and performance of this EULA will be governed by the laws of the Commonwealth of Massachusetts, without regard to conflicts of laws principles.
-
Entire Agreement. This EULA sets forth all of the covenants, provisions, agreements, conditions, and understandings between the parties regarding the subject matter herein, and there are no covenants, promises, agreements, conditions, or understandings, either oral or written, between them other than those set forth herein.
Should you have any concerns regarding this EULA contact us at licensing@edilytics.com.
Cite
For more on how CRISPResso works read the freely available published paper here.
If you like CRISPResso please support us by citing it in your work:
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, Pinello L.
CRISPResso2 provides accurate and rapid genome editing sequence analysis.
Nat Biotechnol. 2019 Mar; 37(3):224-226. doi: 10.1038/s41587-019-0032-3. PubMed PMID: 30809026.
@article{clement2019crispresso2,
title={CRISPResso2 provides accurate and rapid genome editing sequence analysis},
author={Clement, Kendell and Rees, Holly and Canver, Matthew C and Gehrke, Jason M and Farouni, Rick and Hsu, Jonathan Y and Cole, Mitchel A and Liu, David R and Joung, J Keith and Bauer, Daniel E and others},
journal={Nature biotechnology},
volume={37},
number={3},
pages={224--226},
year={2019},
publisher={Nature Publishing Group US New York}
}
Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, Yuan GC.
Analyzing CRISPR genome-editing experiments with CRISPResso.
Nature biotechnology. 2016 Jul;34(7):695-7.
@article{pinello2016analyzing,
title={Analyzing CRISPR genome-editing experiments with CRISPResso},
author={Pinello, Luca and Canver, Matthew C and Hoban, Megan D and Orkin, Stuart H and Kohn, Donald B and Bauer, Daniel E and Yuan, Guo-Cheng},
journal={Nature biotechnology},
volume={34},
number={7},
pages={695--697},
year={2016},
publisher={Nature Publishing Group US New York}
}
CRISPResso Documentation
Select a version below: