About
CRISPResso is a software pipeline designed to enable rapid and intuitive interpretation of genome editing experiments. A limited web implementation is available at: http://crispresso2.pinellolab.org/ or http://crispresso.com.
Briefly, CRISPResso:
- Aligns sequencing reads to a reference sequence
- Quantifies insertions, mutations and deletions to determine whether a read is modified or unmodified by genome editing
- Summarizes editing results in intuitive plots and datasets
Tools
CRISPResso is a suite of complementary tools:
- CRISPResso - for analyzing and interpreting single experimental conditions on a single amplicon
- CRISPRessoBatch - for analyzing and comparing multiple experimental conditions at the same site
- CRISPRessoPooled - for analyzing multiple amplicons from a pooled amplicon sequencing experiment
- CRISPRessoWGS - for analyzing specific sites in whole-genome sequencing samples
- CRISPRessoCompare - for comparing editing between two samples (e.g., treated vs control)
- CRISPRessoAggregate - for aggregating results from previously-run CRISPResso analyses
How can you use CRISPResso?
CRISPResso can be used to analyze genome editing outcomes using cleaving nucleases (e.g. Cas9 or Cpf1) or noncleaving nucleases (e.g. base editors). The following operations can be automatically performed:
- Filtering of low-quality reads
- Adapter trimming
- Alignment of reads to one or multiple reference sequences (in the case of multiple alleles)
- Quantification of HDR and NHEJ outcomes (if the HDR sequence is provided)
- Quantification frameshift/inframe mutations and identification affected splice sites (if an exon sequence is provided)
- Visualization of the indel distribution and position (for cleaving nucleases)
- Visualization of distribution and position of substitutions (for base editors)
- Visualization of alleles and their frequencies
CRISPResso processing
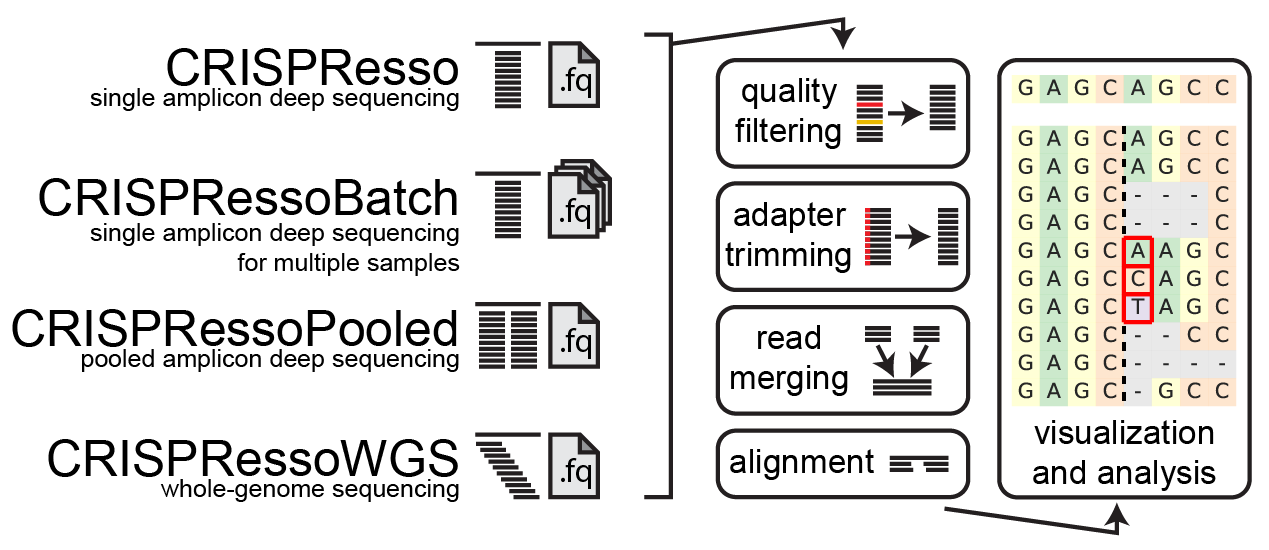
Quality filtering
Input reads are first filtered based on the quality score (phred33) in order to remove potentially false positive indels. The filtering based on the phred33 quality score can be modulated by adjusting the optimal parameters (see additional notes below).
Adapter trimming
Next, adapters are trimmed from the reads. If no adapter are present, select 'No Trimming' under the 'Trimming adapter' heading in the optional parameters. If reads contain adapter sequences that need to be trimmed, select the adapters used for trimming under the ‘Trimming adapter’ heading in the optional parameters. Possible adapters include Nextera PE, TruSeq3 PE, TruSeq3 SE, TruSeq2 PE, and TruSeq2 SE. The adapters are trimmed from the reads using fastp.
Read merging
If paired-end reads are provided, reads are merged using fastp. This produces a single read for alignment to the amplicon sequence, and reduces sequencing errors that may be present at the end of sequencing reads.
Alignment
The preprocessed reads are then aligned to the reference sequence with a global sequence alignment algorithm that takes into account our biological knowledge of nuclease function. If multiple alleles are present at the editing site, each allele can be passed to CRISPResso and sequenced reads will be assigned to the reference sequence or origin.
Visualization and analysis
Finally, after analyzing the aligned reads, a set of informative graphs are generated, allowing for the quantification and visualization of the position and type of outcomes within the amplicon sequence.
How is CRISPResso2 different from CRISPResso?
CRISPResso2 introduces four key innovations for the analysis of genome editing data:
- Comprehensive analysis of sequencing data from base editors. We have added additional analysis and visualization capabilities especially for experiments using base editors.
- Allele specific quantification of heterozygous references. If the targeted editing region has more than one allele, reads arising from each allele can be deconvoluted.
- A novel biologically-informed alignment algorithm. This algorithm incorporates knowledge about the mutations produced by gene editing tools to create more biologically-likely alignments.
- Ultra-fast processing time.